Reticulate - Leveraging Python from R
Python and R, besties forever...
Data Science Altitude for This Article: Camp One.
If you’re a Python developer that has been thrust into an R working environment or an R developer that would like to try out Python packages and methods in the comfort of an already-familiar RStudio IDE, then Reticulate is the package for you. Using rmarkdown, you’re able to knit R and Python code blocks into a unified whole and can refer to objects across the language barrier.
Graphing can not only be done through base plot() or ggplot() calls, but with a little effort we’ll show how to leverage your Python visualization skills using any of the matplotlib, seaborn, or (one of my favorites) bokeh packages.
Further, you have the opportunity to invoke an interactive Python session from within RStudio. How fun is that???
Let’s get to it…
Environment and Prerequisites
RStudio
First, let’s talk about setup. At the time of this article, I’m running RStudio 1.2.1555. I’ve jumped ahead from the publicly-available 1.2.1335 due to some interactions with another project and its use of Pandoc, but 1335 should be good to go for this purpose, it’s been in general availability since early April…
R and Reticulate
I have reticulate 1.13 installed running under an R 3.6.0 base. According to the reticulate documentation page:
SystemRequirements: Python (>= 2.7.0)
Depends: R (>= 3.0)
Imports: utils, graphics, jsonlite, Rcpp (>= 0.12.7), Matrix
Suggests: testthat, knitr, callr, rmarkdown
Python
So, regardless of whether you’re on the Python 2 or 3 fork (a story in itself), you’re safe as Python 2.7 was released back in 2010, and any of the Python 3 releases are sufficient. Having Anaconda on my machine, I’ll be referencing Python and its packages from there. If you use another tool and package consolidator, your code will need to look a little different but you’ll still have the same tasks to do…
Other Packages
I’d highly recommend getting - at a minimum - the knitr and rmarkdown packages updated before you dive in. If you’re not familiar with rmarkdown, I’d highly recommend covering this really good cheatsheet first. A lot of what you’ll see going forward in this post will make a lot more sense. If you’re familiar with HTML markdown syntax, you’ve got a good head-start. Knitr takes the Rmarkdown file and ‘knits’ HTML, PDF, or MS Word documents. If you stray away from HTML, you might have another couple of packages to install. A discussion for another day…
And double-check your version of Rcpp, the package that helps R mesh well with C++ code running behind the scenes. The required 0.12.7 release is quite a while back, but it wouldn’t hurt to check if you don’t update packages frequently…
The rmarkdown setup block
But first, a little discussion of the rmarkdown framework and knitting in RStudio as it relates to reticulate.
```{r setup, warning = FALSE}
knitr::opts_chunk$set(echo = TRUE)
library(reticulate)
use_python("D:/Anaconda3") # Path to python3.exe
```Setup should be your initial rmarkdown block, and it has some special connotations accompanying it. Suffice it to say that it’s preferable to have all your library invocations and up-front work stationed there. After specifying the reticulate library, the important inclusion is the path to your machine’s python3.exe. Or, if you’re using Python 2, to that specific .exe file. For me, it’s in my Anaconda library.
Before that, though, is the lowercase ‘r’ that notes that the block is using the r engine. If that leaves you spinning, please reference the rmarkdown cheatsheet linked earlier as well as material in the ‘Further Information’ section at the bottom of the post.
Here, the warning = FALSE suppresses awareness of which version of R that reticulate was built under. It might be good to know initially, but necessarily not over repeated builds…
Our First Python Chunk
Our initial entry within the curly braces shows that this chunk should use the Python engine. The syntax in the remainder of the block should look rather familiar to Python developers and leverages the pandas package, home of Python’s Source and DataFrame data structure implementations. It’s part of the sciPy group of packages that all practicing open-source Data Scientists should be (or become…) familiar with.
You can find a copy of the flights.csv data file here, if interested. We’re taking a look at flights where the flight distance is less than 1800 miles. We’ll slim it down and look at just the carrier field and the arrival and departure differentials (both early and late) from the stated flight time, along with the distance flown. Nothing too strenuous, but enough to illustrate some of the reticulate’s functionality for cross-chunk data reference.
```{python}
import pandas as pd
flights = pd.read_csv("flights.csv")
flights = flights[flights['distance'] < 1800]
flights = flights[['carrier', 'dep_delay', 'arr_delay', 'distance']]
flights = flights.dropna()
print(flights.head())
```## carrier dep_delay arr_delay distance
## 0 UA 2.0 11.0 1400
## 1 UA 4.0 20.0 1416
## 2 AA 2.0 33.0 1089
## 3 DL -6.0 -25.0 762
## 4 UA -4.0 12.0 719Referencing Python structures in an R chunk
To reference Python variables in an R chunk, you have to preface them with ‘py$’:
```{r}
sprintf("%d of %d flights were delayed more than fifteen minutes.",
sum(py$flights$dep_delay > 15), length(py$flights$dep_delay))
```## [1] "20221 of 118284 flights were delayed more than fifteen minutes."Also, you can throw ggplot at them using they ‘py$’ reference, if your visualization skills are better with ggplot than the Python renderers:
```{r}
library(ggplot2)
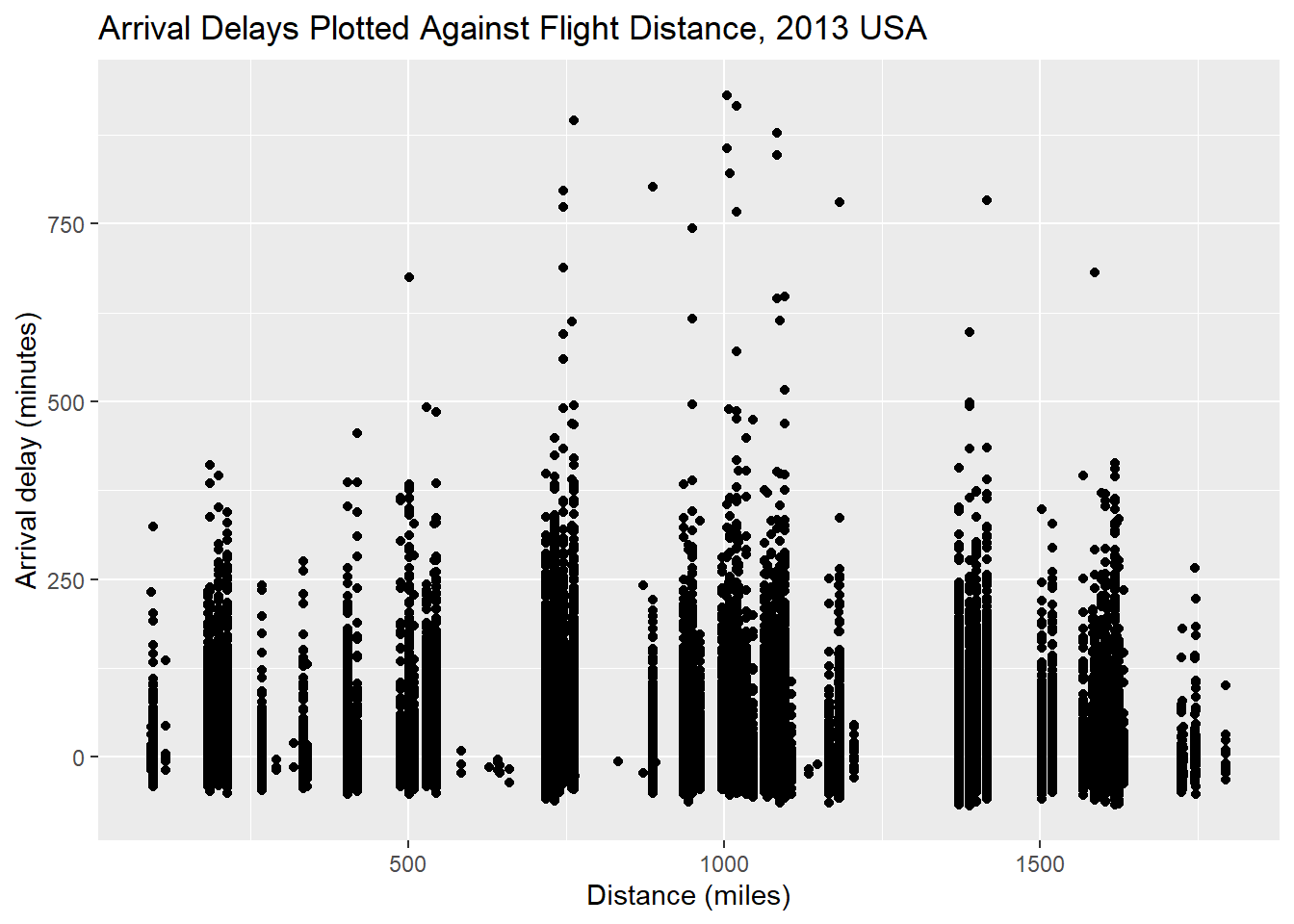
p <- ggplot(py$flights, aes(y = arr_delay, x = distance)) + geom_point()
p + labs(x = 'Distance (miles)', y = 'Arrival delay (minutes)',
title = 'Arrival Delays Plotted Against Flight Distance, 2013 USA')
```
Let’s take a look at what it takes in a Python chunk to refer to R data…
Referencing R structures in Python.
So, let’s reference a few variables from the python data that only exist in an R chunk. Below is the code to create the mean departure and arrival delays of the subset greater when zero. If you’re going to depart late or arrive late, what’s the average time without the influence of the times you were early?
```{r, collapse = TRUE}
meanDepDelay = mean(py$flights$dep_delay[py$flights$dep_delay > 0])
meanArrDelay = mean(py$flights$arr_delay[py$flights$arr_delay > 0])
```You can do that in a Python block by using r.objectName:
```{python, collapse = TRUE}
print(r.meanDepDelay)
print(r.meanArrDelay)
```## 34.518
## 36.381Okay, so far so good. We can refer to R data in Python, and vice versa. But what if you’re wanting to roll the other direction - you’ve got r variables but are more comfortable throwing a Pythonic visualization out there? No problem, but there’s an extra step involved…
Using Python renderers within an rmarkdown block.
There’s one thing you need to do before trying a piece of code like this:
```{python, collapse = TRUE}
from math import cos, pi
import numpy as np
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.pyplot as plt
from matplotlib import cm
xDomain = list(np.arange(-5.12, 5.12, .08))
yDomain = list(np.arange(-5.12, 5.12, .08))
X, Y = np.meshgrid(xDomain, yDomain)
z = [20 + x**2 + y**2 - (10*(cos(2*pi*x) + cos(2*pi*y))) for x in xDomain for y in yDomain]
Z = np.array(z).reshape(128,128)
fig = plt.figure(figsize = (12,10))
ax = fig.gca(projection='3d')
surf = ax.plot_surface(X, Y, Z, cmap=cm.RdYlGn, linewidth=1, antialiased=False)
ax.set_xlim(-5.12, 5.12)
ax.set_ylim(-5.12, 5.12)
ax.set_zlim(0, 80)
fig.colorbar(surf, aspect=30)
plt.title('Rastrigin Function in Two Dimensions: (Min at 0, Max at 80)\n 20 + x^2 + y^2 - 10*cos(2*pi*x) - 10*cos(2*pi*y)\n')
plt.show()
```Looks good syntactically, but it will blow up on you.
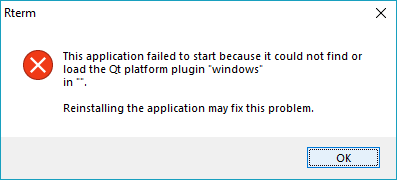
You’ll first need to set an environment variable for the PyQt cross-platform enablement package, bridging the C++ package Qt and Python. Again, Anaconda to the rescue:
import os
os.environ['QT_QPA_PLATFORM_PLUGIN_PATH'] = 'D:\Anaconda3\Library\plugins\platforms'Once you add in those lines before the plt.show() call, you’re good to go.
## (-5.12, 5.12)
## (-5.12, 5.12)
## (0, 80)
## <matplotlib.colorbar.Colorbar object at 0x000000001E1F3828>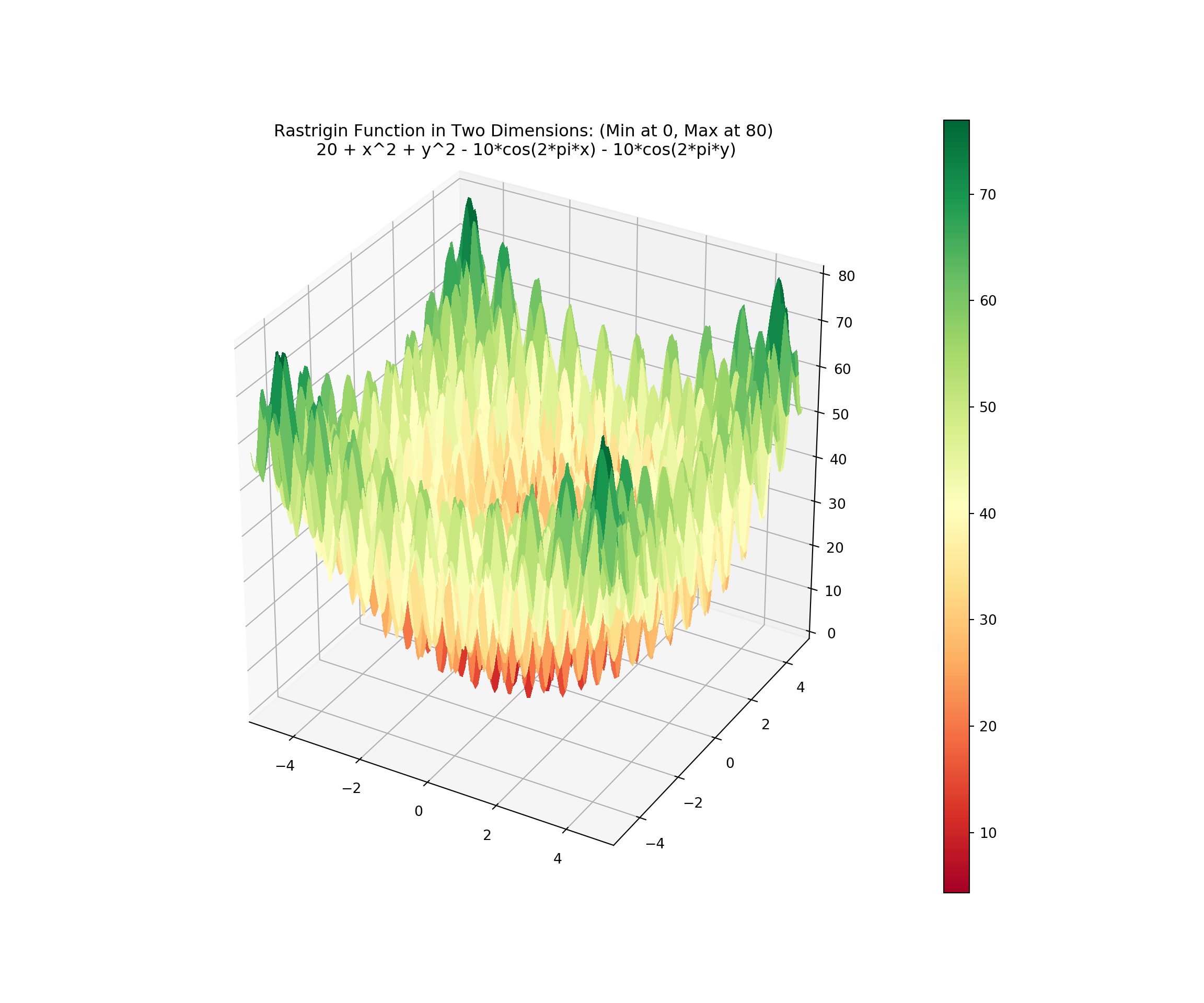
An Interactive Python Environment in the R Console
Running an interactive Python session in the R console is as simple as:
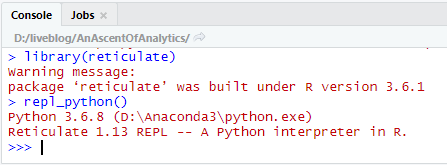
Note that it uses the version of python in our earlier use_python() call. From here, let’s try out some code that uses the Symbolic Python package sympy:
> repl_python()
Python 3.6.8 (D:\Anaconda3\python.exe)
Reticulate 1.13 REPL -- A Python interpreter in R.
>>> from sympy import Symbol, plot, Derivative, pprint, solve, Circle, Point
>>> from sympy.plotting import plot_parametric
>>> a,b,c,d,low,high = (-1.5, -12.3, 6, 500, -10, 10)
>>> x = Symbol('x')
>>> polyCubic = a*x**3 + b*x**2 + c*x + d
>>>
>>> # The smallest y-value for positioning the axes of the plot doesn't necessarily
>>> # have to happen at the low or high of the x ranges.
>>> yExtremes = (polyCubic.subs(x, high), polyCubic.subs(x, low))
>>>
>>> p = plot(polyCubic, (x, low, high), axis_center = (low, min(yExtremes)),
... title = "y-values for {0}x^3 {1:+}x^2 {2:+}x {3:+} between {4} and {5}".format(a,b,c,d,low,high),
... show = False)
>>> # Determine derivative of equation and if roots are non-complex and can be plotted
>>> dx =Derivative(polyCubic, x)
>>> dxSoln = dx.doit()
>>> print("Derivative of Cubic equation is:")
Derivative of Cubic equation is:
>>> pprint(dxSoln, use_unicode = True)
2
- 4.5⋅x - 24.6⋅x + 6
>>>
>>> roots = solve(dxSoln)
>>> if isinstance(roots, complex):
... print("Roots of the derivative have components in the complex plane (a + bi).")
... else:
... point1 = polyCubic.subs(x, roots[0])
... point2 = polyCubic.subs(x, roots[1])
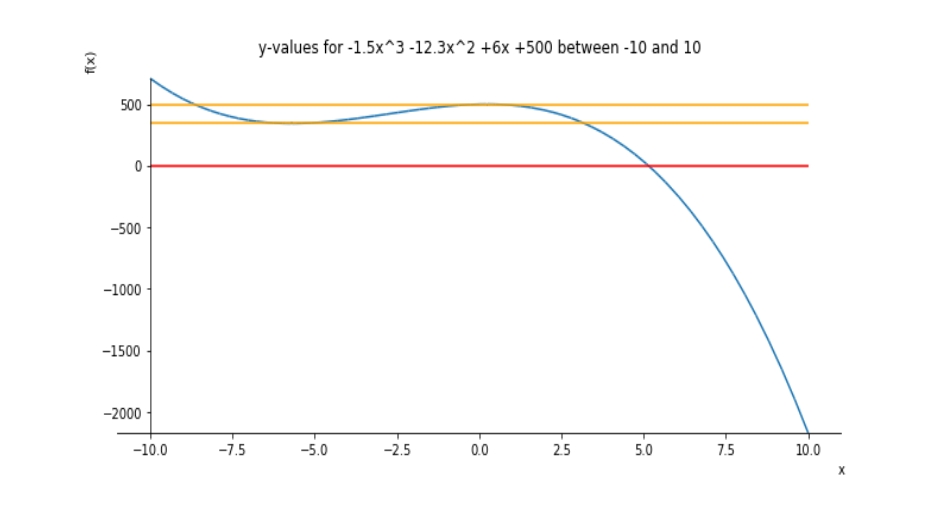
>>> print("The real roots of the cubic derivative are x = {0:.3f} and x = {1:.3f}".format(roots[0], roots[1]))
The real roots of the cubic derivative are x = -5.701 and x = 0.234
>>> p2 = plot_parametric(x, 0, line_color = 'red', show = False)
>>> p.append(p2[0])
>>> p3 = plot_parametric(x, point1, line_color = 'orange', show = False)
>>> p.append(p3[0])
>>> p4 = plot_parametric(x, point2, line_color = 'orange', show = False)
>>> p.append(p4[0])
>>> p.show()You should get something like this as an output:
Let’s also try sourcing something from the command line. Here’s the output from a piece of python code from the bokeh gallery.
source_python('bokeh_source.py')You should get something that looks like this in a separate HTML window:
Not much more to say, please take a look at all the documentation and enjoy !
Further Information on the Subject:
RMarkdown, explained. Kudos to Yihui Xie, J.J. Allaire, and Garrett Grolemund.
Reticluate pages: From RStudio and from Pablo Franco.
